After isolating the bacteria from two different kind of fish mucus, the first important test to bacteria identification is antibiotic sensitivity test. Antibiotic sensitivity is a test where you test the marine bacteria against other kind of bacteria that lives in different environment, has different host, from different species and etc that cause diseases. Thus, we can test marine bacteria against human pathogens or any other kind of pathogens.
As mentioned before, this experiment, antibiotic sensitivity will take four days to complete.
Objective: To determine the presence of the presence of antibiotic sensitivity in microbe.
Materials:
Unsterilized: 8 kind of human pathogens, 14 petri dishes (with marine nutrient agar), parafilm, hockey stick, pipette, ampicillin
Sterilized: centrifuge tubes, tips, disk filter paper in glass petri dish, nutrient broth/agar in schott bottle, scraper, flasks
Method-
Day 1
1. Prepare all cell suspension.
2. Prepare 7 centrifuge tubes flled with 5ml of nutrient broth.
3. Inoculate the pathogens into each nutrient broth.
4. shake in incubator 30C, 200 rpm, 24 hours.
For marine bacteria suspension (grow again the bacteria of 11 and 33 from the flasks into new flasks):
1. Inoculate the bacteria into 5ml marine nutrient broth.
2. Shake in incubator 30C, 200 rpm, 24 hours.
Day 2
1. Clean the laminar flow and get it ready for the next experiment.
2. Materials needed are 14 petri dishes and labeled with the names of the seven pathogens with two petri dishes for each pathogen and also divided the petri dishes into four compartments.
3. First, spread the petri dishes with pathogens on the agar surfaces by using hockey stick.
4. After that, put the disk paper into four different compartments, with each different solution for each compartment- C is for control, Amp is for ampicillin, 11 is for bacteria 11 and 33 is for bacteria 33. Do the same for the rest of the petri dishes.
5. Store the petri dishes in fridge 2-4 C for 24 hours (pre-diffusion techniques)
Day 3
Take the petri dishes out and store in ambient temperature.
Day 4
Observe result
Result:
The eight human pathogen been tested with are:
1. Enterobacter Aerogenes
2. Pseudomonas Aeruginosa
3. Staphylococcus Aerus
4. Streptococcus Faecalis
5. Serfatia Marcesceus
7. Enterococcus Feacalis
8. E. Coli
9. Bacillus Subtilis
(Noticed that number 6 is missing as there was a mistake in labeling)
Result:
The pictures below shows the result from testing the marine bacteria with the different kind of human bacteria. There were six bacteria been chosen from all the marine bacteria been grow. They are S1aI, K1AII, S1AII, K1BII, K1AV and S1BIV.
For each of the Petri dish it is divided into four different compartments, with C as control, in which the small round filter paper is not dip into any solution; Amp stands for ampicillin (in which the round filter paper is been dipped into ampicillin solution), and the rest of the two compartments represent any of the marine bacteria listed above. The positive result is determined by a hollow zone (empty sphere) surrounding the small round disk filter paper.
No 1, Enterobacter Aerogeues

K1BII and S1AII

K1AV and S1BIV
Picture for S1AI and K1AII is blur but there is no hollow zone found.
No 2, Pseudomonas Aeruginosa

S1AII and K1BII

K1AV and S1BIV

S1AI and K1AII
No 3, Staphylococcus Aerus

K1BII and S1AII

K1AV and S1BIV

S1AI and K1AII
No 4, Staphylococcus Feacilis

K1BIi and S1AII

S1AI and K1AII
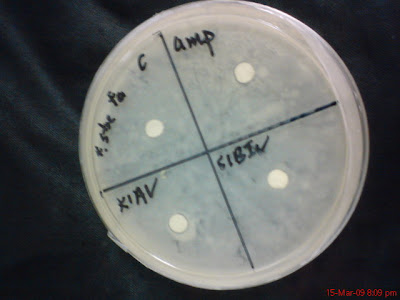
K1AV and S1BIV
No 5, Serfatia Marcesceus

K1BII and S1AII

K1AV and S1B IV

K1BII and S1AII
No 7, Enterococcus Feacalis
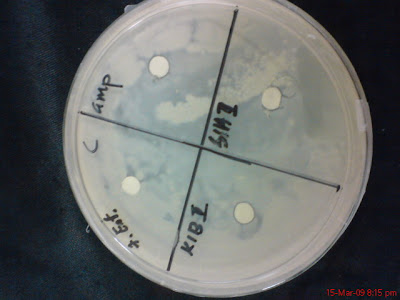
K1BII and S1AII

S1AI and K1AII

K1AV and S1BIV
No 8, E. Coli
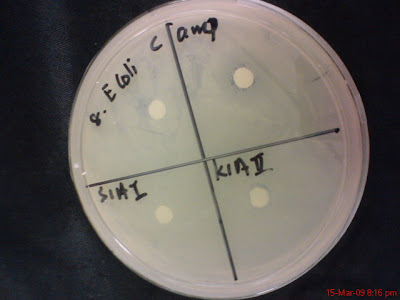
S1AI and K1AII
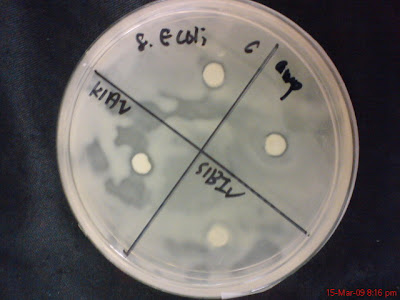
K1AV and S1BIV

KlB II and S1AII
No 9, Bacillus Subtilis

K1BII and S1AII

S1AI and K1AII
Picture for bacteria K1BII and K1BV is blur but there is also no hollow zone found.
Out of 24 plates only one plate show possibility of hollow zone and that is the plate with bacteria S1AI and K1AII with No 3 Staphylococcus Aerus pathogen, like below.

Because I didn't observe the growth of the bacteria in the plates from time to time but instead only after 24 hours, so it is suspected that this plate has hollow zone but its just that the bacteria (K1AII and S1AI) grow over the area that has been cleared. This plate will be repeat both in Marine Nutrient Agar and as well as Distilled water Nutrient Agar.
Conclusion:
One plate will be repeated in two different medium to reconfirm the result.